Next-Generation Sequencing (NGS) was introduced to the market about fifteen years ago, as a faster way to do DNA sequencing than first-generation Sanger technology. A lot faster, in fact: while Sanger sequencing can read about 1,000 - 2,000 DNA bases in one individual reaction, one NGS run yields up to 6 billion.
NGS didn't replace Sanger sequencing, but its higher speed and power made DNA sequencing available for many new applications. One of the companies taking full advantage of its potential is Eurofins Genomics. In April of this year, the company announced that in November 2018 it started to provide NGS services on the NovaSeq6000, the latest and most updated sequencing platform developed by Illumina.
rapidmicrobiology asked Dr. Thomas Brefort, Business Unit Manager for NGS at Eurofins Genomics, and one of the Managing Directors of Eurofins' sequencing division, about the impact the NovaSeq6000 had on their NGS services, and what the future of this technology looks like.
Q. What type of clients do you typically offer your NGS services to and for what applications?
A. We offer our next-generation sequencing to quite a wide range of clients, like global pharma, biotechnology and agricultural technology companies, as well as academic institutions.
Whole Genome Sequencing is, of course, one of the applications of NGS, but our portfolio of services is not limited to that.
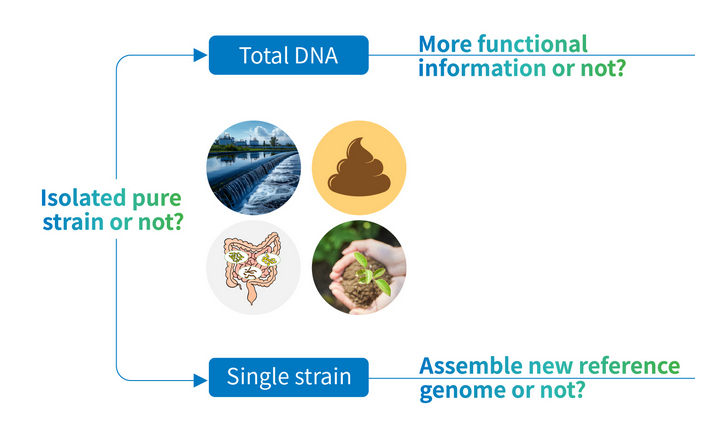
We perform microbiome or bacterial analysis, which can be used in different scenarios, for example, to monitor or assess skin microbiome in order to identify diseases or to help plant breeders know what beneficial or harmful microbiota live in and on their plants.
There is also a growing interest from the food industry, where NGS can exactly identify foodborne pathogens and trace them back to their sources, or to prevent food fraud by making sure that - for example - what is sold as tuna is not a cheaper type of fish.
In the clinical field, we offer exome sequencing for cancer research, not only on tumour tissues but also on liquid biopsies, which use a simple blood draw from patients to identify actionable genome mutations. We have years of experience with liquid biopsy approaches for whole exomes and oncopanel.
Q. How has the NovaSeq6000 expanded your range of services?
A. It is allowing us to sequence DNA five times faster than before, with a lower cost per base. Additionally, it works with all kinds of input material, like degraded samples. That had been a limitation of some of Illumina’s previous equipment, which was initially restricted to human sample material.
We have a much higher throughput now, and that means we can really scale our capacity to manage more and even bigger projects in less time, adjusting the outputs to our needs. Increased speed allows us to offer faster turnaround times, which is very important in foodborne illness outbreak scenarios and in food manufacturing for example. The cheaper cost per base also allows us to be more competitive in the human genome sequencing area, where you need large amounts of sequencing data.
Q. How was the new machine implemented?
A. Before installing the NovaSeq6000 last year, we had done our homework and worked a lot on the preparation. As a result, implementation was seamless.
First, we designed our own set of unique dual indexes (UDIs) to resolve index hopping, which is a typical issue of Illumina´s latest NGS flowcells and can lead to cross-contamination between samples. Our Eurofins Genomics Oligonucleotide Synthesis colleagues synthesized these sets, which were then thoroughly qualified by NGS before applying them for our routine production. These NGS-approved sets of Eurofins UDIs will shortly also be available as a separate oligonucleotide product.
After the installation, our R&D team ran a comprehensive series of tests to evaluate and compare results for every different pipeline and documented the whole technical validation procedure. Tests confirmed that output and quality with the NovaSeq6000 were even significantly better than before.
Q. Where is NGS headed to in the next five to ten years?
A. I think that what used to be a highly sophisticated technology will be more and more adopted for everyday applications, whether it’s diagnostics, medicine, food, agriculture or direct-to-consumer testing. These growing demands and opportunities are also already reflected in our growing portfolio. NGS is able to provide answers to challenges which we previously couldn't address and this will benefit the broader public.
In order to support this wider adoption and acceptance of NGS, the direction is to simplify handling of the equipment as well as protocols for library preparation. In fact, that is something we’ve done internally, by combining different processing pipelines into lean and simple automated handling processes.
Additional advanced sequencing technologies are emerging, called third-generation sequencing, with the potential for generating longer reads and being more accurate, but they do not have the throughput of second-generation technology.
As DNA sequencing becomes faster, more scalable and more automated, however, it will be important to make sure that results are still reliable. That is achieved using a dedicated LIMS system that tracks samples from their arrival throughout the whole process.
Further resources
Here below are five recent publications with findings of international research studies in four different areas, where Eurofins Genomic's R&D team participated.
As Dr. Brefort explained: "Studies like these are a great way for us to drive innovation in general as well as internally. They also showcase the quality of our scientific approach and know-how and the comprehensiveness of our portfolio."
1. Microbiome: Association between the cervicovaginal microbiome, BRCA1 mutation status, and risk of ovarian cancer: a case-control study.
2. Exome sequencing: Optimised molecular genetic diagnostics of Fanconi anaemia by whole-exome sequencing and functional studies.
3. Epigenetic markers in cell-free cancer DNA: The potential of circulating tumor DNA methylation analysis for the early detection and management of ovarian cancer.
4. Methylation patterns in serum DNA for early identification of disseminated breast cancer.
5. Sequencing and analysis of bacteria: Discovery of monoclonal antibodies cross-reactive to novel subserotypes of K. pneumoniae O3
Find out more about Eurofins Genomics NGS services