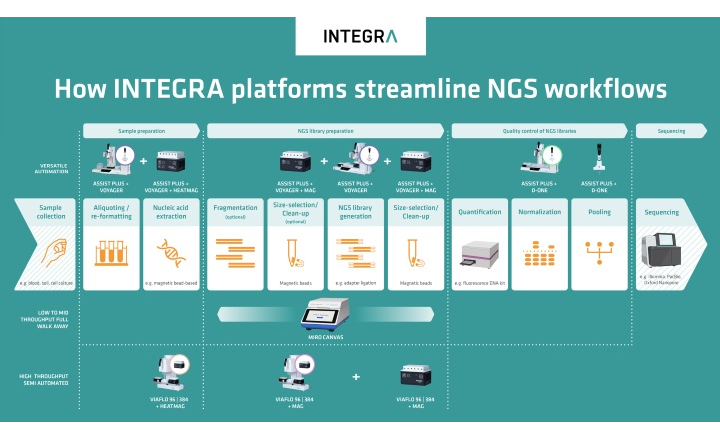
Oxford Nanopore Technologies and NVIDIA are collaborating this year to integrate the NVIDIA DGX Station A100 into Oxford Nanopore’s ultra-high-throughput sequencing system, PromethION.
Partnering the NVIDIA A100 Tensor Core GPU technology with the PromethION device aims to deliver the world’s most powerful sequencer that supports real-time analyses at scale, and can also analyse any length fragment of DNA/RNA.
The use of accelerated computing and artificial intelligence to quickly and accurately sequence DNA or RNA supports the increasing availability of nanopore sequencing data, at scale, to a variety of high-throughput users. Oxford Nanopore’s technology is increasingly being used by scientific researchers analysing many thousands of genomes to better understand genetic diversity and discover new variants. Sequencing is also being increasingly adopted to generate rapid insights in healthcare settings, food safety, or environmental analysis.
NVIDIA DGX Station A100, announced in November, is a data-centre-grade, GPU-powered, multi-user workgroup appliance that can tackle the most complex AI workloads. It plugs directly into an outlet in an office or laboratory and is very quiet thanks to its refrigerant-based cooling system. It contains four NVIDIA A100 80GB GPUs, fully connected via NVIDIA NVLink, to offer a total of 320GB of GPU memory.
Unprecedented analytical power for ultra-high-throughput sequencing
The 2.5 petaFLOP AI compute system from NVIDIA offers unprecedented compute density, performance and flexibility in a benchtop format. Oxford Nanopore’s PromethION P48 sequencing device continues to challenge even the most powerful devices with its ability to generate as much as 10 Terabases of DNA information per 72-hour run (sufficient to analyse 96 human genomes at 30X coverage).
Breaking through the 10 Terabase run barrier was announced by Oxford Nanopore at the Community meeting in December 2020 and represents a 25% increase in data output compared to its previous best earlier in the same year. This increase has been driven by continual improvements in flow cell chemistry, many of which were included in new shipments from mid-November 2020. These developments have been reflected in customer data, with increasing yields reported across a range of sequencing applications.
Supplied in a P24 and a P48 format, PromethION is increasingly being deployed into high-throughput projects, where the rich sequencing data provided by Oxford Nanopore can be delivered at very high throughput. As with all Oxford Nanopore devices, the technology enables academic groups, core facilities and service providers to realise the value of sequencing any length fragments, from short fragments to those that are over 100,000 bases long, and to characterise base modifications, coupled with high accuracy single nucleotide or structural variant calling and phasing.
NVIDIA GPUs are already used in other Oxford Nanopore sequencing systems, driving real-time sequencing analysis at any scale. The desktop GridION includes NVIDIA V100 technology and the handheld MinION Mk1C sequencer is powered by the NVIDIA Jetson Edge AI platform.
AI, training and algorithm development as a driver of high accuracy sequence data
The use of powerful AI systems is also driving substantial improvements in the accuracy of Oxford Nanopore’s sequencing data; updated analysis algorithms can result in higher accuracy of the same sequence data.
Oxford Nanopore recently released a new machine-learning driven analysis algorithm, Bonito CRF, with which users have reported >98% single read basecalling accuracy. Basecalling is the process of identifying the sequence of bases on an individual molecule of DNA. This latest update to Bonito builds on previous work to deliver improved performance and is trained with a larger, more diverse data set.
High single read accuracy supports very high consensus accuracy (sequencing multiple times for higher accuracy); optimised analysis tools including Guppy/Bonito basecalling, assembly with Canu/Flye and polishing with Medaka, can now enable Q45 with R9.4.1 flow cells and Q50 with R10.3 flow cells.
Variant calling performance is also improving with the latest releases. Using the latest tools, structural variation (SV) accuracy has reached gold standard of 96% with 30X rather than 60X coverage. Oxford Nanopore has now seen SNV at 99.92%, which is comparable to traditional SBS accuracy.
In addition, in late 2020 Oxford Nanopore generated modal single-read accuracy of 99.1% (99%=Q20) using a new chemistry with Bonito, delivered on internal validation sets, with a substantial proportion of these raw reads above Q20.
Oxford Nanopore and NVIDIA are working closely to deploy the latest advancements in AI, with the goal of making biological analysis available to anyone, anywhere.
Note: This content has been edited by a rapidmicrobiology staff writer for style and content.